AllerCatPro 2.0
Contents
2. Input and output of AllerCatPro 2.0 3. Profilin proteins 4. Very large proteins 5. Nucleotide input 6. Benchmark datasets 7. Browser compatibility 8. Conceptualizers and contacts 9. Consultant 1. About AllerCatPro 2.0
Due to the risk of inducing an immediate Type I (IgE-mediated) allergic response, proteins intended for use in consumer products must be investigated for their allergenic potential before introduction into the marketplace. Therefore, it is crucial to have proteomic/bioinformatic tools to accurately predict and investigate allergenic potential of proteins. In this study, we have developed our AllerCatPro 2.0 web server for comprehensive analysis and prediction of allergenicity potential from the protein/nucleotide sequence, and visualization of 3D models for the input protein based on the similarity of 3D surface epitopes. AllerCatPro 2.0 provides a user-friendly interface to identify protein allergenicity potential with detailed results for cross-reactivity, protein information (UniProt/NCBI), functionality (Pfam, InterPro, SUPFAM), as well as clinical relevance of IgE prevalence (Allergome) and allergen information of the most similar allergen.
We define “allergenicity potential” or “allergenic potential” as the potential of a protein to cause/elicit immediate-type (IgE-mediated) allergic reactions in humans (Blackburn et al., Crit Rev Toxicol., 2015). We are not able (yet) to accurately predict sensitizing from non-sensitizing (but potentially cross-reactive) allergens (Krutz et al., Crit Rev Toxicol., 2020). Therefore, we would consider “allergenicity potential” as the ability to sensitize an individual and/or potentially cross-react in pre-sensitized individuals. In the absence of methods to characterize the relative allergenic potential of proteins, the default assumption judges all proteins to be potentially allergenic unless there is evidence for the contrary. Nevertheless, it seems inappropriate to designate proteins as non-allergens in the absence of robust methods to identify the inherent lack of allergenic potential. However, individual proteins with a significant relative abundance in protein sources for which there are known to be significant opportunities for human exposure, but no evidence for allergenicity, can be considered with low allergenic potential. This paradigm to allow the identification of allergenic proteins that display low sensitizing potential has been proposed by Krutz et al. (Krutz et al., Toxicol Sci., 2019).
Gluten-like Q-repeats are identified using the method in our previous study (Maurer-Stroh et al., Bioinformatics, 2019). From the FARRP AllergenOnline database, “Celiac disease peptides” are downloaded in our dataset. The amino acid frequencies are calculated for every 9-mer window within the peptides and a composition fingerprint score is computed by using a log odd ratio of the frequency in the “Celiac 9-mer” windows divided by a background database frequency (UniProtKB). This log odd score is used to score all 9-mers in a query sequence and it triggers a hit as Gluten-like Q-repeat if the score for a 9-mer is within one standard deviation of the average of the FARRP “Celiac disease peptides”.
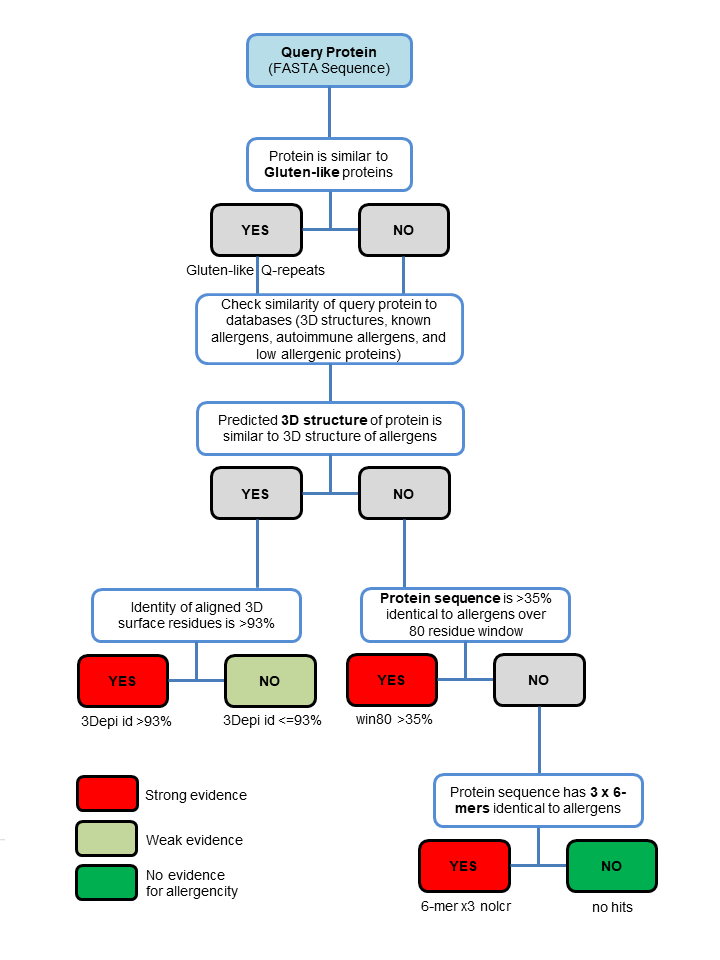
Decision workflow of AllerCatPro 2.0 from the query protein to the results of either strong, weak or no evidence for allergenic potential. AllerCatPro 2.0 checks the similarity of the query protein with 714 representatives in our 3D model/structure database of known allergens as well as the most comprehensive dataset of reliable proteins associated with allergenicity (4979 protein allergens). In addition to only comparing the similarity of the query protein with the dataset of known allergens in AllerCatPro 1.7, AllerCatPro 2.0 now predicts the similarity of the query sequence to datasets of 165 autoimmune allergens and 162 low allergenic proteins separately. If a significant sequence similarity is found, then AllerCatPro 2.0 identifies hits of similar proteins associated with autoimmune diseases and/or similar proteins of low allergenic potential and presents the sequence identity to the closest hit.
2. Input and output of AllerCatPro 2.0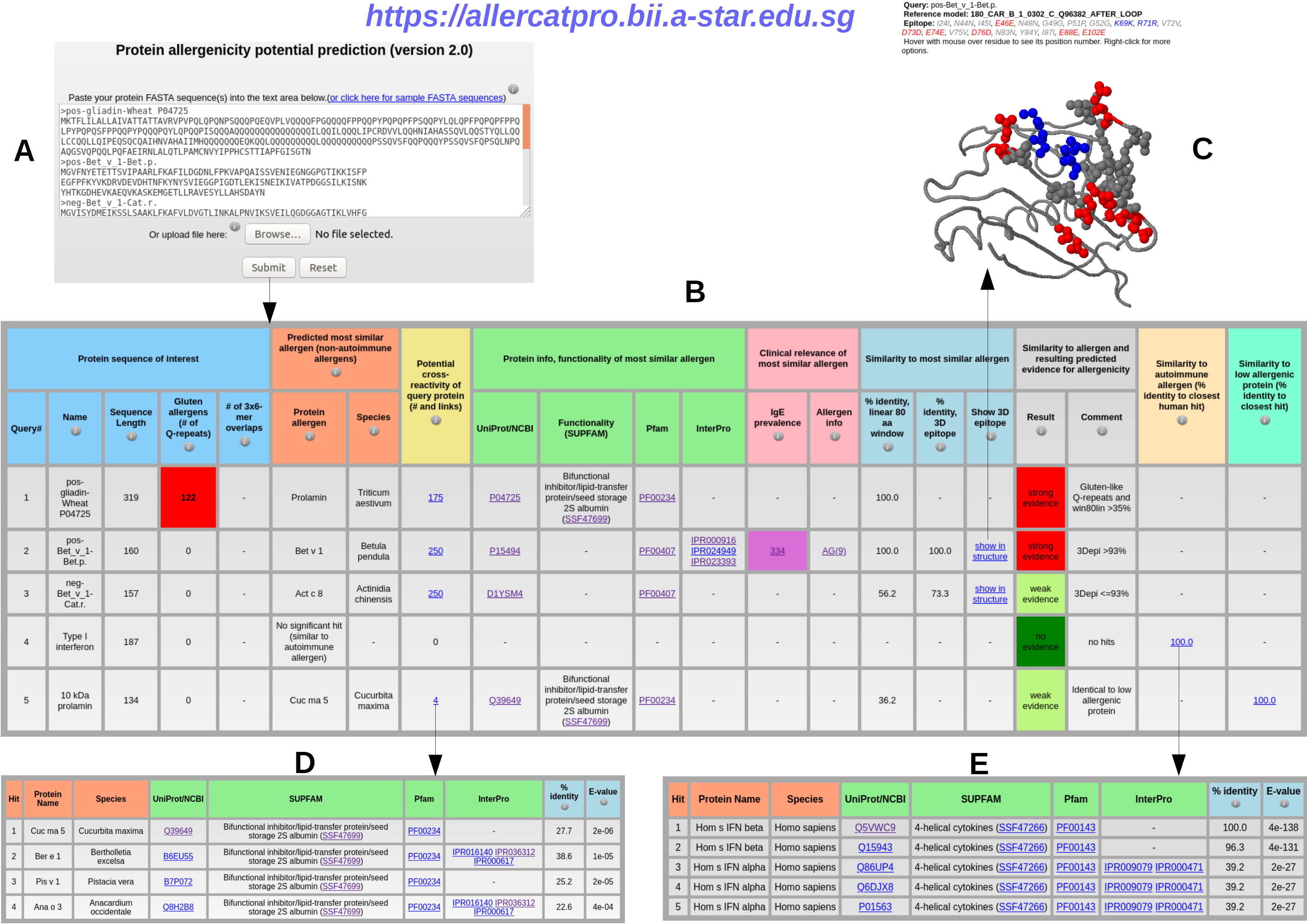
Input: submitting one or more protein sequences in FASTA format (A) leads to AllerCatPro 2.0 output table with the result for strong, weak or no evidence for allergenicity per protein based on corresponding workflow decisions and, in case of a hit, the possibility to view the most similar allergens with detailed results for cross-reactivity, protein information (UniProt/NCBI), functionality (Pfam, InterPro, SUPFAM), as well as clinical relevance of IgE prevalence (Allergome) and allergen information (B), the most similar 3D surface epitope via links with the structural view showing identical epitope residues as balls colored as blue for positive charges, red for negative charges and gray for all other amino acid types (C). AllerCatPro 2.0 also identifies all similar allergens that have significant sequence similarity to the query protein and refers to the number with the link in potential cross-reactivity of the output table (D), as well as all possible similar autoimmune allergens displayed in the link (E) and all possible similar low allergens in the link of the output table.
The detailed help information and explanation of input sequence, uploaded file and output results are provided when user hovers with mouse over their 'information' buttons 3. Profilin proteinsProfilin proteins play relevant roles as confounding factors as well as sensitizer in both diagnosis and treatment for patients with plant food and pollen allergy (Rodríguez Del Río et al., J Investig Allergol Clin Immunol, 2018). In this case study, we analyse and predict allergenicity potential of different profilin proteins (allergenic profilins from pollen and plant as well as low allergenic profilins from yeast, human, cow, chicken, and fungus) using AllerCatPro 2.0 Input: The dataset of pollen profilins of European white birch, olive, timothy, rice from Betula pendula, Olea europaea, Phleum pratense, Oryza sativa is available at the following link Pollen_profilin.fa 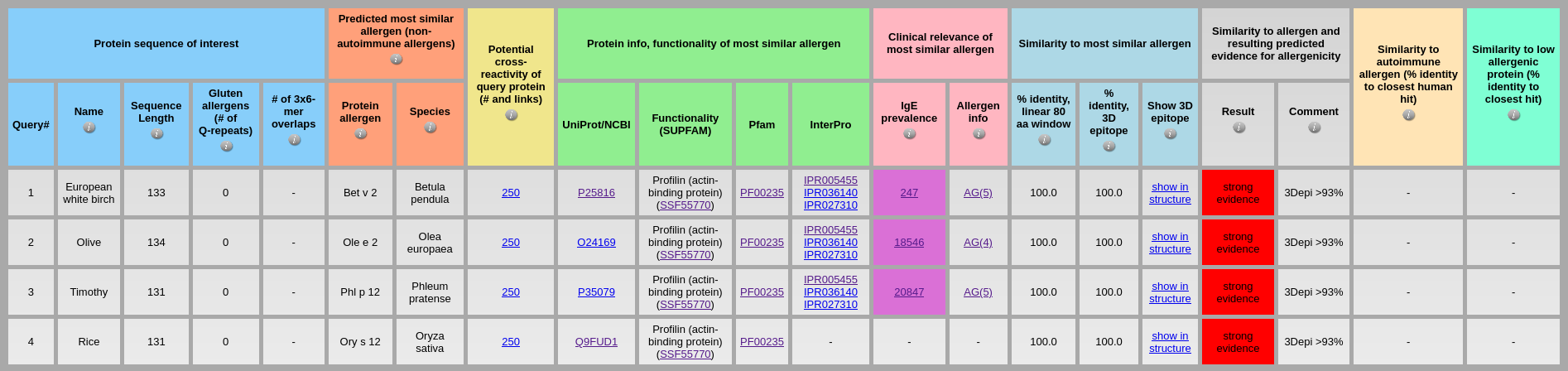
Output: The results of AllerCatPro 2.0 in the above output table show that all pollen profilins of European white birch, olive, timothy, rice are classified correctly as allergenic proteins with ‘strong evidence’. In adition, the predicted of most similar allergens for pollen profilins of European white birch, olive, and timothy have significantly high number of IgE prevalence. The functionality of most similar allergen from SUPFAM also indicates that these proteins are profilins (actin-binding protein). Input: The dataset of plant profilins of peach, apple, hazelnut, potato, spinach, and Para rubber tree is available at the following link Plant_profilin.fa 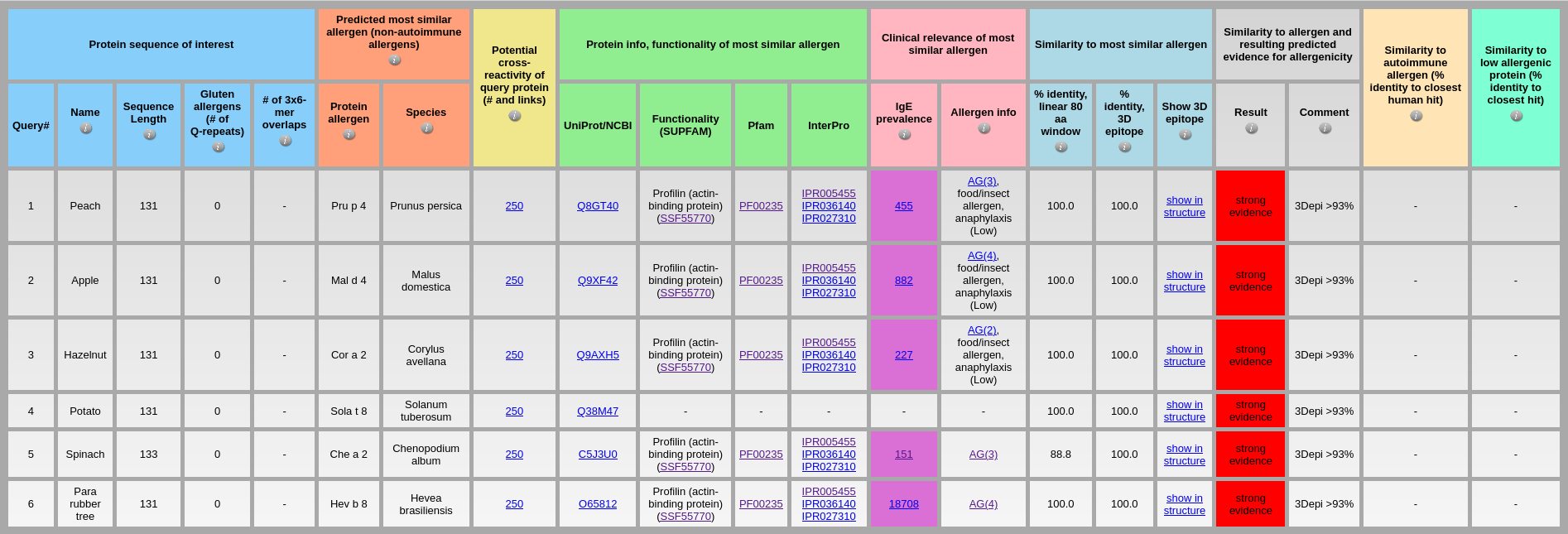
Output: As shown in the above output table, AllerCatPro 2.0 classifies correctly as allergenic proteins with ‘strong evidence’ for all these plant profilins. The predicted of most similar allergens for plant profilins from peach, apple, hazelnut, spinach, and Para rubber tree have significantly high number of IgE prevalence. Input: The dataset of low allergenic profilins from yeast, human, cow, chicken, and fungus is available at Profilin_low_allergens.fa 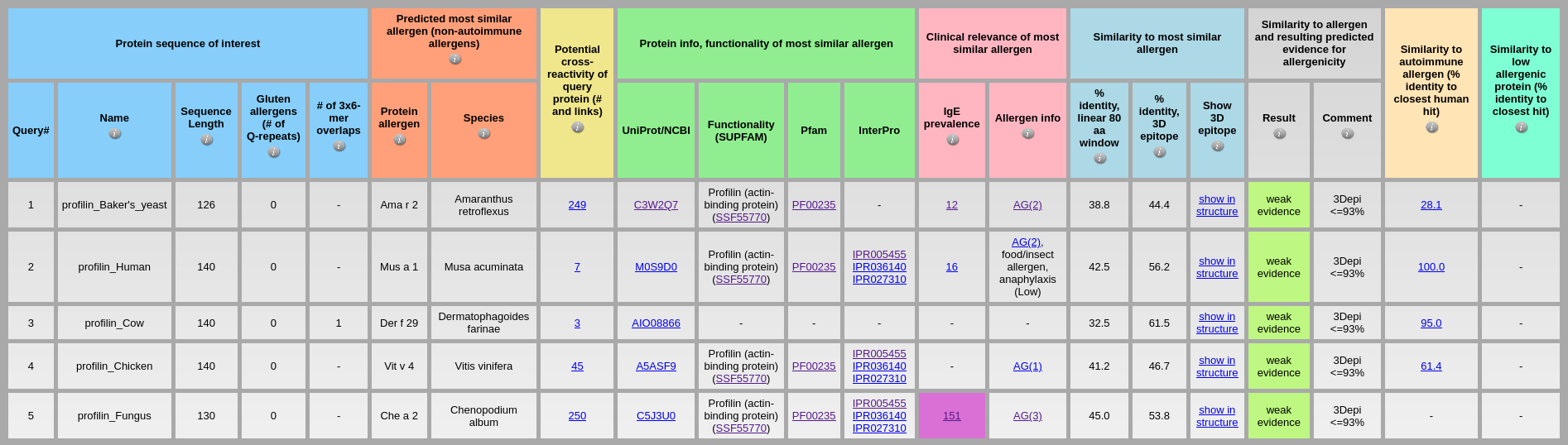
Output: The results of AllerCatPro 2.0 show that all low allergenic profilins from yeast, human, cow, chicken, and fungus are predicted correctly for their allergenicity potential with ‘weak evidence’ prediction.
4. Very large proteins
One of limitations of AllerCatPro is that it could not perform well for very large proteins (>1000 amino acids). In AllerCatPro 2.0, we have improved our method to predict protein allergenicity potential for very long input sequences.
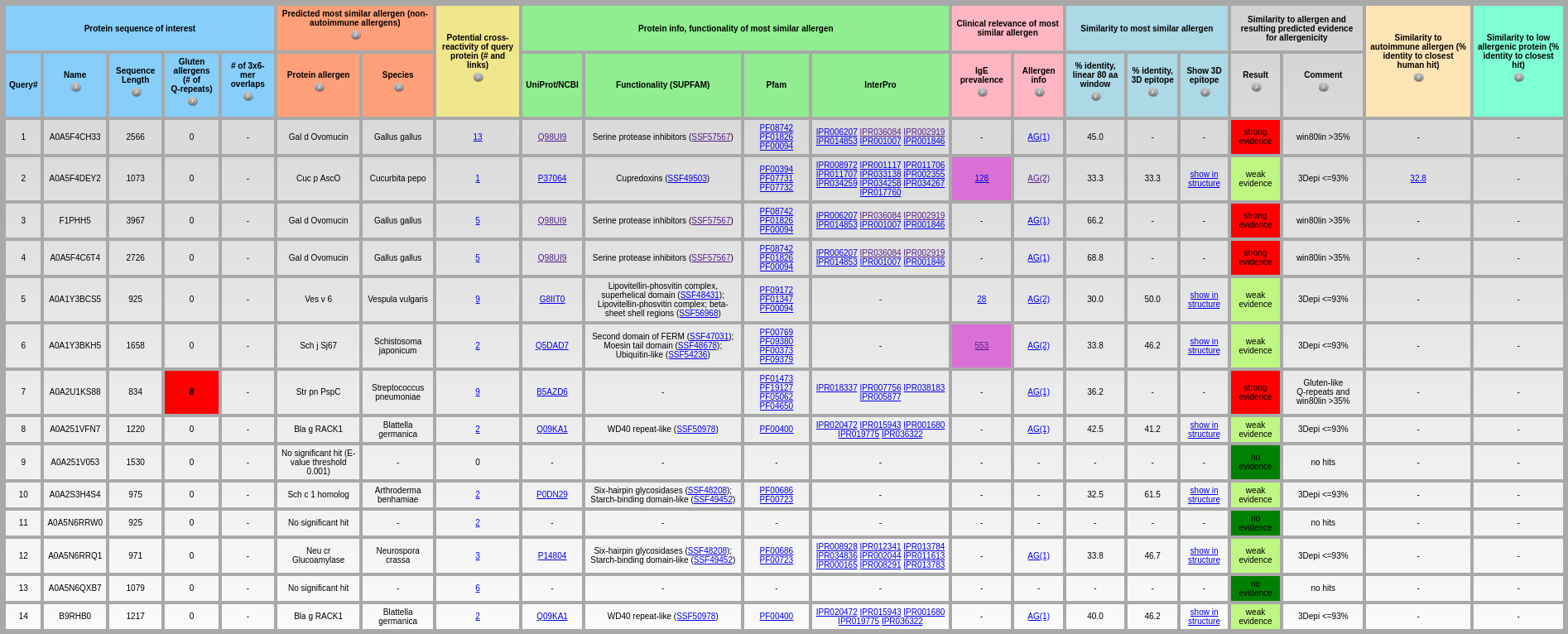
Output: The above output table shows the results of AllerCatPro 2.0 for predicting allergenicity potential for the dataset of very large proteins. The average running time for each protein in this dataset is approximately 8.5 seconds. On this dataset, the allergenicity potential of input protein sequence is identified using different levels of similarity to known allergens in our datasets based on Gluten-like Q-repeat, 3D epitopes, linear-window rule.
5. Nucleotide input
In addition, we have extended the AllerCatPro method to allow users to input nucleotide sequences in AllerCatPro 2.0
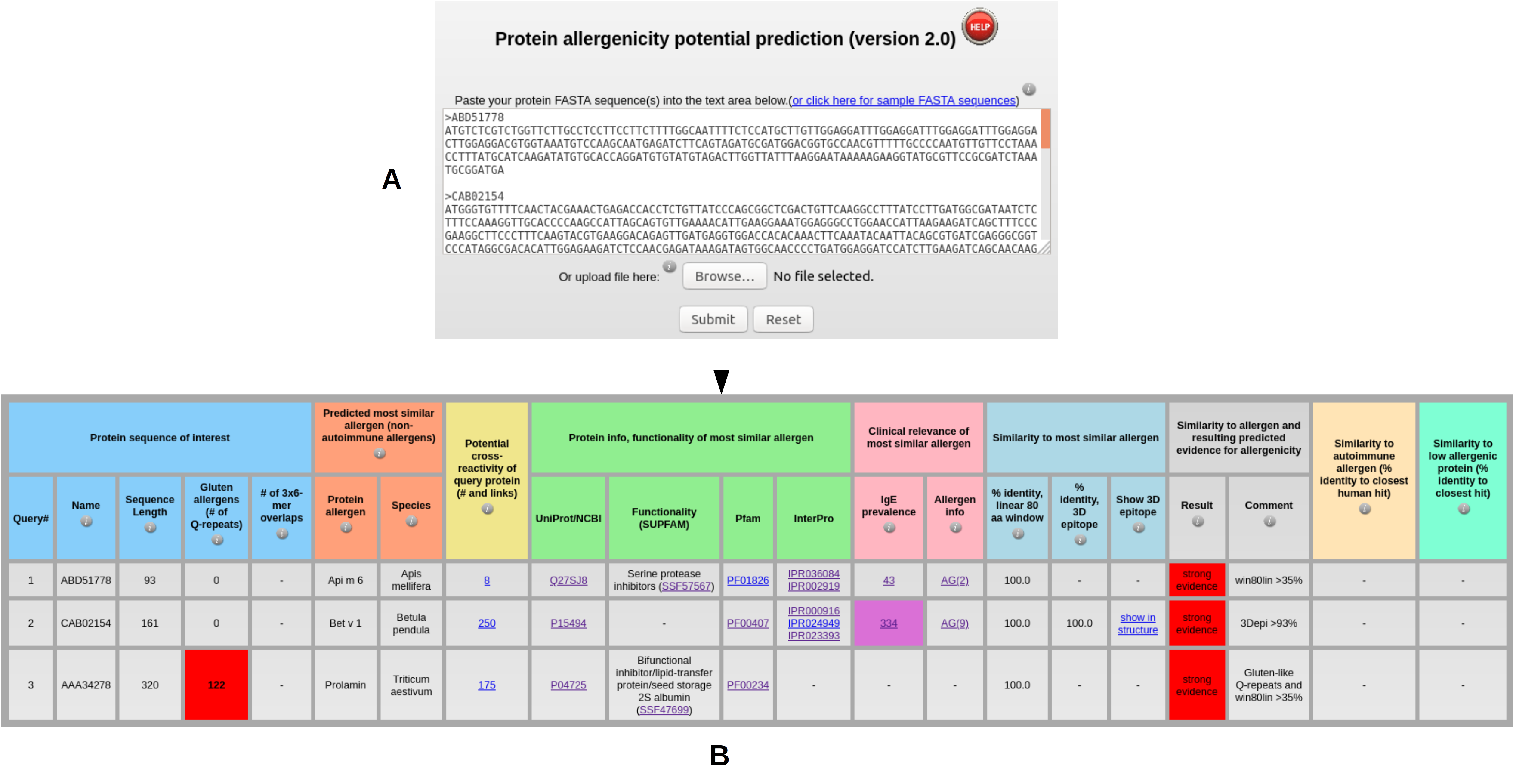
Output: The output table (B) shows the results of AllerCatPro 2.0 for predicting allergenicity potential for the dataset of nucleotide sequences from Apis mellifera, Betula pendula, and Triticum aestivum (A). AllerCatPro 2.0 predicts all these input sequences as allergens with ‘strong evidence’.
6. Benchmark datasets
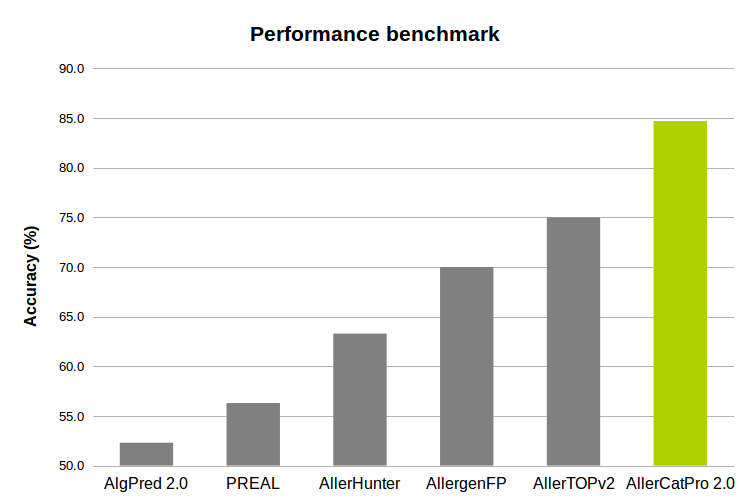
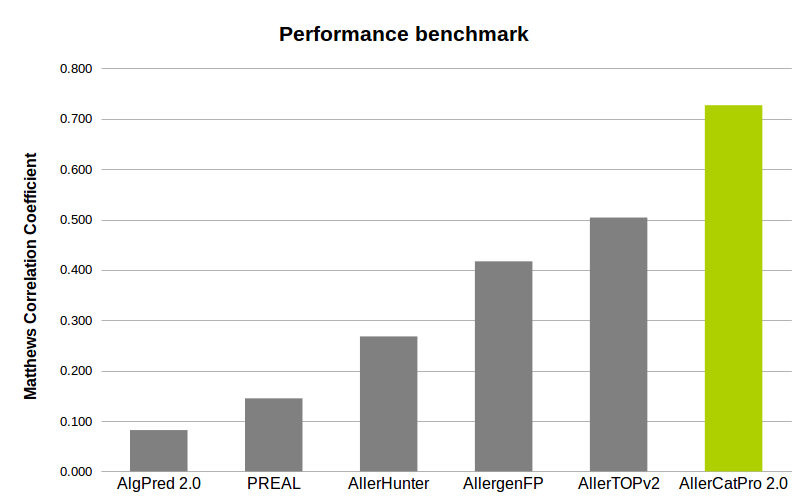
AllerCatPro 2.0 is compared to other methods on the benchmark datasets of 218 positive (known allergen) and 212 negative (likely non-allergen) sequences. These datasets are extracted from the test sets of 221 positive and 221 negative sequences (Maurer-Stroh et al., Bioinformatics, 2019) after we carefully checked and removed one low allergen (‘Spi o RuBisCo’ of Spinacia oleracea) and two autoimmune allergens in the positive set, and nine known allergens ('Ory s 14' of Oryza sativa) in the negative set as these 'Ory s 14' proteins have recently shown evidence of allergenicity in Allergome. This is difficult benchmark because of allergens versus non-allergens with same structure fold (Maurer-Stroh et al., Bioinformatics, 2019).
The accuracy (A) and Matthews correlation coefficient (B) of PREAL, AllerHunter, AllergenFP, AllerTOPv2, AlgPred 2.0 and AllerCatPro 2.0 on the benchmark datasets of 218 allergens and 212 non-allergens with the same structure fold.
There are 1132 sequences of protein allergens, low allergenic proteins, and autoimmune allergens newly included into the datasets of AllerCatPro 2.0.
On these sequences, AllerCatPro 2.0 achieves accuracy of 99.5%.
We perform AllerCatPro 2.0 on the larger validation datasets of 2003 positive (allergen) and 2015 negative (non-allergen) sequences from AlgPred 2.0 (Sharma et al., Briefings in Bioinformatics, 2021). These sets are extracted from the validation datasets of 2015 positive and 2015 negative sequences of AlgPred 2.0 (https://webs.iiitd.edu.in/raghava/algpred2/stand.html) with the following adaptations: We removed one protein (P_10228) that is identical to low allergenic protein (Phoenix dactylifera) and eleven proteins (P_7089, P_7095, P_7102, P_7098, P_7101, P_7108, P_7125, P_7117, P_7114, P_7092, P_7111) associated with autoimmune diseases in the positive set.
We test AllerCatPro 2.0 on all recent IUIS allergens (twenty proteins) that has been created/modified from December 2021 to April 2022. For these new IUIS allergens, AllerCatPro 2.0 predicts their allergenicity potential correctly for fourteen proteins with ‘strong evidence’, five proteins with 'weak evidence’, and incorrectly for only one protein with 'no evidence'.
7. Browser compatibility
8. Conceptualizers and contacts
A*STAR: Minh N. Nguyen, Vachiranee Limviphuvadh, and Sebastian Maurer-Stroh*
9. Consultant
We have a consultancy package if users and/or industries need more guidance on results from AllerCatPro. Please contact A*STAR directly: allercatpro@bii.a-star.edu.sg
|